|

Fig. 1.

Fig. 2.

Fig. 3.
|
Cotton leaf curl geminivirus
(CLCuV) causes a major disease of cotton in Asia and Africa. Leaves of
infected cotton curl upward (Fig. 1) and bear
leaf-like enations on the underside along with vein thickening (Fig.
2). Plants infected early in the season are stunted and yield is reduced
drastically. Severe epidemics of CLCuV have occurred in Pakistan in the
past few years, with yield losses as high as 100% in fields where infection
occurred early in the growing season.
Another cotton geminivirus, cotton leaf crumple virus
(CLCrV), occurs in Arizona, California, and Mexico. CLCrV symptoms are
distinguishable from CLCuV symptoms in that infected leaves curl downward
accompanied by interveinal hypertrophy and foliar mosaic (Fig.
3).
Both CLCrV and CLCuV infect dicotelydonous plants and
are whitefly-transmitted (Brown et al., 1983; Mansor et al., 1993). Previous
studies (Brown and Nelson, 1984; 1987; Hameed et al., 1994; Mansor et al.,
1993) suggested that they belong to the subgroup III geminiviruses. However,
little information is available on the relationship of these two viruses
with each other and with other subgroup III geminiviruses.
|
|
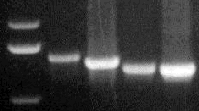
PCR-amplified DNA fragments from
both CLCrV and CLCuV were subsequently sequenced. Analysis of nucleic acid
sequences indicates that CLCuV is closely related to tomato yellow leaf
curl virus, cassava latent virus, and Indian cassava mosaic virus from
the old world, whereas CLCrV is more closely related to potato yellow mosaic
virus, bean dwarf mosaic virus, and bean golden mosaic virus from the New
World.
Together, these data clearly suggest
that CLCrV occuring in North America and CLCuV occuring in Asia and Africa
are two distinct viruses.
|