Resources
Wasp collecting
We collect wasps using standard Drosophila traps, which can be set up in any number of ways (see Drosophila: A Guide To
Species Identification And Use). Any trap that attracts fruit flies is also likely to attract parasitoid
wasps looking for Drosophila
hosts. Usually we use large plastic storage containers, with slits on the sides for fly entry, that
are filled with rotting fruit.
Having a sealable lid is great to prevent your trap from getting rained on and to prevent varmints
from making off with the
fruit. The wasps are small (~2mm) and look like mini ants with wings, and they can often be found
exploring the inside of
rotting fruits for fruit fly larvae and pupae. Unlike flies, wasps rarely fly when the traps are opened and
are thus caught in nets
infrequently. Instead, after netting off the flies from the trap, we catch the wasps by aspirating them
off the substrate into
standard Drosophila food vials for storage. To rear the wasps we catch (see below), we put them in
vials with early-instar fly
larvae from whatever Drosophilids were attracted to the trap. We later try to move the new wasps
onto one of the easy-to-rear
and generally susceptible fly species, like D. melanogaster or D. virilis.



Wasp rearing
Drosophila parasitoid wasps are pretty easy to rear in the lab. If you can rear Drosophila, you can rear their parasitoid wasps.
For larval parasitoids, we allow host flies to lay eggs in standard Drosophila food vials for two to three days until there are a
good number (>100) of fly eggs. We then remove the adult flies and replace them with adult wasps, using at least 5 female
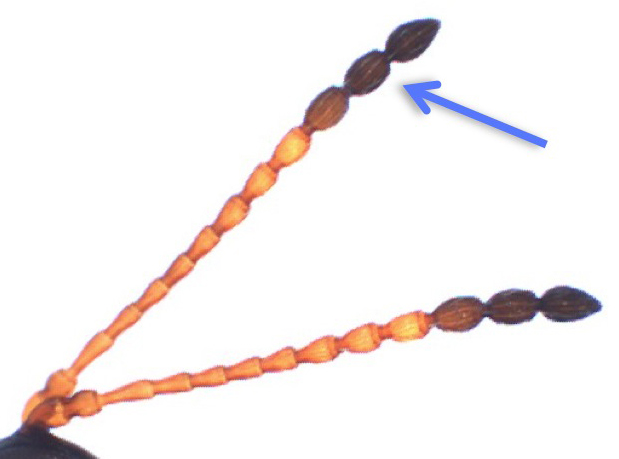
wasps per vial. Figitid females can be distinguished from males by their much shorter antennae, while Braconid females have
obvious ovipositor sheaths projecting from their abdomens. The wasps usually attack the second instar fly larvae and will
complete their life cycles and eclose in 3-4 weeks at room temperature. For pupal parasitoids, we wait until fly larvae are at
wandering stage before adding wasps. Diapriid females can be distinguished from males by their much shorter antennae.
We've found that adding ~0.5mL of 50% honey (by volume) in water to the cotton tops of the Drosophila vials prolongs wasp
lifespan. Wasps can be treated just like flies, e.g. they can be knocked out with CO2 and can be maintained alive for long
periods of time at 18 degrees C.

Wasp identification
We use Hymenoptera Of The World: An Identification Guide To Families to determine which
families new wasps belong to.
For identification of Figitid wasps in the genus Leptopilina, we've found four papers
particularly useful (Nordlander 1980,
Allemand et al. 2002, Novkovic et al. 2011, Lue et al. 2016). Keys for other
Drosophila parasitoids tend to be obscure and
limited to particular species. We are developing a morphological key for the
wasps we have, but sometimes it's easiest to
collect DNA sequence information for wasp species
identification and compare it to relevant databases like Genbank or the
more specific DROP database (Lue et al. 2021). We
obtained
COI sequence for all our wasps as well as ITS2 sequence for
the Figitids using the following PCR primers, although
more recent primer sets may be found in Lue et al. 2021.
COI forward: ggtcaacaaatcataaagatattgg
COI reverse: taaacttcagggtgaccaaaaaatca
ITS2 forward: tgtgaactgcaggacacatg
ITS2 reverse: aatgcttaaatttagggggta
Genbank numbers for all sequences are listed on the phylogeny below, but you can download all our sequences in one shot
here. If you happen to catch some Drosophila parasitoids and aren't sure what they are, we would be happy to try identifying them
for you and possibly add them to our stock collection!

Wasp strains
We maintain a large number of live strains of parasitoid wasps that infect Drosophila. Our lab policy is to make these strains
available to everyone once we have published on them (subject to the conditions of the relevant USDA permits that we hold).
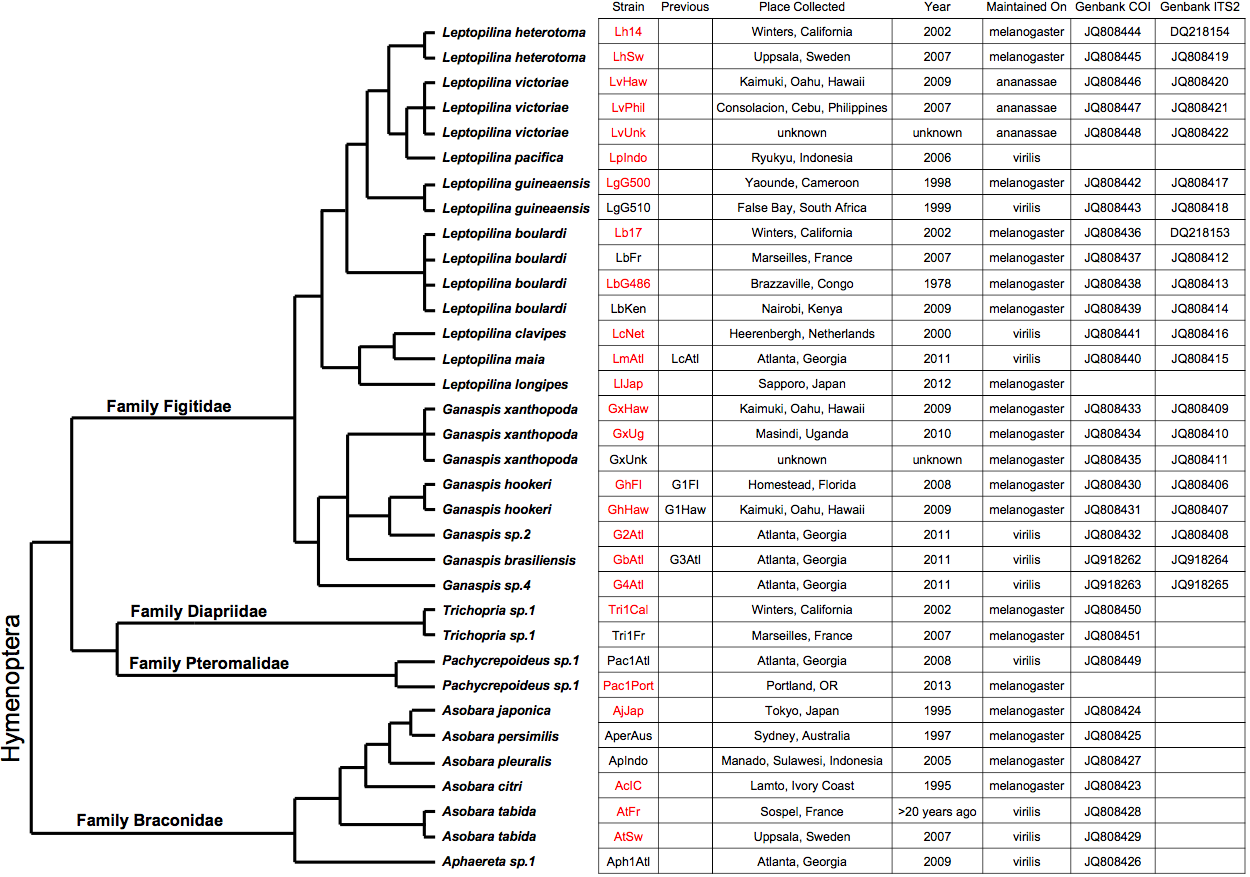
The phylogeny below shows all the strains we've grown in the past, many of which became available with our publication on
wasp resistance in D. suzukii (Kacsoh and Schlenke 2012). The strain names shown in red are the ones that we currently
grow. Phylogenetic relationships and branch lengths are approximated and are based on Hymenopteran family relationships
from Dowtin and Austin 2001, Figitid relationships from Schilthuizen et al. 2002, Allemand et al. 2002, Novkovic et al. 2011,
and our own work, and Braconid relationships from
Seyahooei et al. 2011 and our own work.
Fly transcriptional response to wasp infection
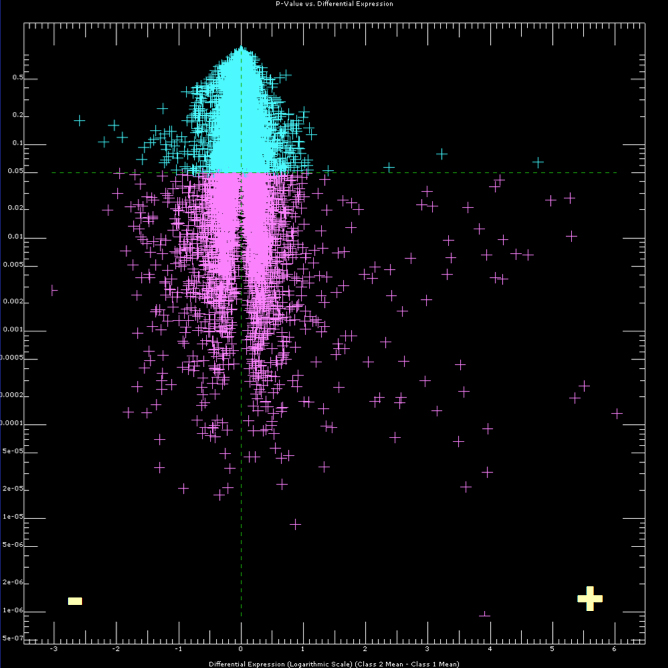
In 2007 we published a microarray study comparing the transcriptional responses of D. melanogaster to two different
parasitoid wasp species (Schlenke et al. 2007). The data were deposited in the Gene Expression Omnibus, accession number
GSE8938. However, for quick access to the data to check if your favorite fly gene was differentially expressed in our study,
please download this excel file of gene expression fold changes. In the file, bold numbers represent statistically significant
fold changes. "Lb-5" corresponds to fold expression change in fly larvae infected by the wasp L. boulardi, compared to
uninfected flies, five hours after infection. Likewise, "Lh-24"corresponds to fold expression change in fly larvae infected by the
wasp L. heterotoma, compared to uninfected flies, twenty four hours after infection. The "LbLh" values combine L. boulardi
and L. heterotoma infection data for comparison against uninfected fly data while the "Lb/Lh" values compare expression
between L. boulardi and L. heterotoma infected flies.
Wasp venom genes
In 2013 we published two papers describing a joint transcriptomic-proteomic approach for identifying parasitic wasp venom
genes (Goecks et al. 2013, Mortimer et al. 2013). The data were deposited in various databases, including in GenBank for
transcriptome data (Transcriptome Shotgun Assembly accession numbers GAJA00000000, GAJC00000000,
GAIW00000000) and the PRIDE database for proteomic data (accession numbers PDX023836, PDX023824, PDX023825).
For quick access to the lists of venom-specific transcripts and protein sequences from the three wasp species, please
download the following Fasta files for the wasps L. boulardi (cDNA, protein), L. heterotoma (cDNA, protein), and Ganaspis
hookeri (cDNA, protein). The cDNA sequences in these files are the female wasp abdomen transcript contigs that were "hit" by
peptide sequences collected from the lumens of female wasp venom glands, while the protein sequences are the translated
ORFs from the wasp transcripts that were hit by the the venom peptides.


